🚀 @SBERLOGABIO webinar on bionformatics and data science:
👨🔬 Robert Hoehndorf, Maxat Kulmanov, "DeepGO-SE - Protein function prediction as approximate semantic entailment"
⌚️ Thursday 6 June, 18.30 (Moscow time). Unusual time - pay attention.
Add to Google Calendar
The talk is based on Nature Machine Intelligence paper: https://www.nature.com/articles/s42256-024-00795-w#Sec1 presenting a beautiful, original approach to the task to predict protein properties (Gene Ontology terms) based on the protein sequence (like CAFA5 Kaggle challenge). From leading experts in the field.
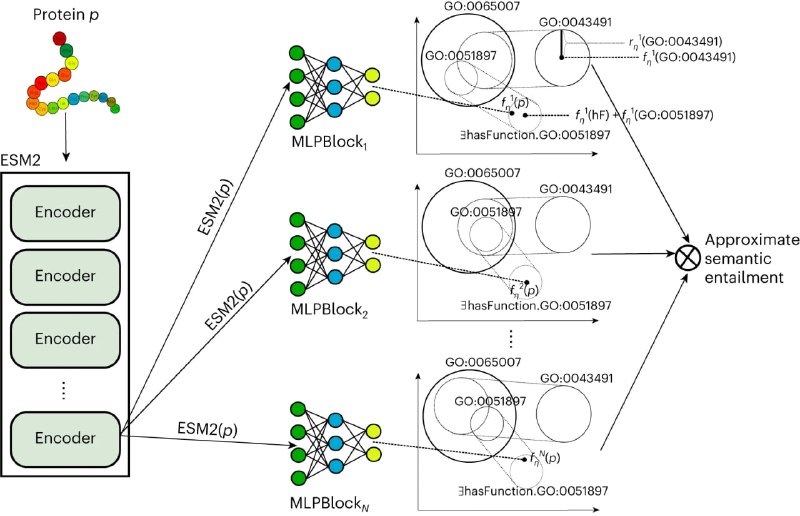
The Gene Ontology (GO) is a formal, axiomatic theory with over 100,000 axioms that describe the molecular functions, biological processes and cellular locations of proteins in three subontologies. Predicting the functions of proteins using the GO requires both learning and reasoning capabilities in order to maintain consistency and exploit the background knowledge in the GO. Many methods have been developed to automatically predict protein functions, but effectively exploiting all the axioms in the GO for knowledge-enhanced learning has remained a challenge.
In this webinar, I will present DeepGO-SE, the latest version of DeepGO methods, that predicts GO functions from protein sequences using a pretrained large language model. DeepGO-SE incorporates the knowledge in GO by learning multiple approximate models of GO using an ontology embedding method. Furthermore, it uses a neural network to predict the truth values of statements about protein functions in these approximate models. We aggregate the truth values over multiple models so that DeepGO-SE approximates semantic entailment when predicting protein functions. We show, using several benchmarks, that the approach effectively exploits background knowledge in the GO and improves protein function prediction compared to state-of-the-art methods.
📹 Video: https://youtu.be/vhnD4SR8cWI?si=000BSzLtjsvc_bvC
📖 Presentation: https://t.me/sberlogabio/74962
👨🔬 Robert Hoehndorf, Maxat Kulmanov, "DeepGO-SE - Protein function prediction as approximate semantic entailment"
⌚️ Thursday 6 June, 18.30 (Moscow time). Unusual time - pay attention.
Add to Google Calendar
The talk is based on Nature Machine Intelligence paper: https://www.nature.com/articles/s42256-024-00795-w#Sec1 presenting a beautiful, original approach to the task to predict protein properties (Gene Ontology terms) based on the protein sequence (like CAFA5 Kaggle challenge). From leading experts in the field.
The Gene Ontology (GO) is a formal, axiomatic theory with over 100,000 axioms that describe the molecular functions, biological processes and cellular locations of proteins in three subontologies. Predicting the functions of proteins using the GO requires both learning and reasoning capabilities in order to maintain consistency and exploit the background knowledge in the GO. Many methods have been developed to automatically predict protein functions, but effectively exploiting all the axioms in the GO for knowledge-enhanced learning has remained a challenge.
In this webinar, I will present DeepGO-SE, the latest version of DeepGO methods, that predicts GO functions from protein sequences using a pretrained large language model. DeepGO-SE incorporates the knowledge in GO by learning multiple approximate models of GO using an ontology embedding method. Furthermore, it uses a neural network to predict the truth values of statements about protein functions in these approximate models. We aggregate the truth values over multiple models so that DeepGO-SE approximates semantic entailment when predicting protein functions. We show, using several benchmarks, that the approach effectively exploits background knowledge in the GO and improves protein function prediction compared to state-of-the-art methods.
📹 Video: https://youtu.be/vhnD4SR8cWI?si=000BSzLtjsvc_bvC
📖 Presentation: https://t.me/sberlogabio/74962